David Becerra
I received a PhD degree in Computer Science with emphasis in Bioinformatics in 2017 from McGill University. During my doctoral studies, I worked under the supervision of Professor Jérôme Waldispühl.
From 2018 to 2020, I was a post-doctoral fellow under the supervision of Professor Philip Kim in the Terrence Donnelly Centre for Cellular & Biomolecular Research.
I recieved my undergrad and master degrees from the Universidad Nacional de Colombia, where I developed my master thesis and undergrad capstone project under the supervision of Professor Luis Niño
I joined the School of CS at McGill in 2020 as a Faculty Lecturer. I really enjoy researching and teaching courses in bioinformatics and algorithms. I am also highly interested in programming competitions.
Teaching at McGill University.
Comp204: Computer Programming for Life Sciences: Fall'21, Fall'22
Comp208: Computer Programming for Physical Sciences: Fall'20
Comp251: Algorithms and Data Structures: Winter'21, Winter'21, Winter'22
Comp321: Programming Challenges: Fall'20, Winter'21, Fall'21, Winter'22, Fall'22
Research.
It focuses on the area of Computational Biology and Algorithms. I am mainly concerned with applying computational techniques to the modeling and simulation of complex problems in Life Sciences. Particularly, related to the use of Machine Learning, Bio-inspired Computing, Data Mining, Algorithms and Optimization Processes in the solution of molecular biology problems at a sequential and structural level.
Programming Competitions
We have witnessed an increase in the number of our students involved in competitive programming teams. Through this participation, students from different backgrounds and coding abilities have earned algorithmic experience as well as confidence in both their computing and interpersonal skills. Furthermore, this collaborative environment has fostered a welcoming and inclusive culture in which students have found a safe space to train and to share their competitive hobby with friends and classmates alike. Contestants have gone on to represent McGill in various international competitions. Among other honors, we qualified for the International Collegiate Programming Contest ( ICPC ) world finals during 2020 and 2021. ICPC boasts over 50,000 student participants representing some 3,000 universities in 111 different countries, all competing for just 100 final spots. Two years in a row, McGill has sent a qualifying team. We always aim to welcome even more talented students to take part in our competitive programming teams - perhaps you will represent us in the next world finals! If you are interested in joining the teams, please contact me.

Publications
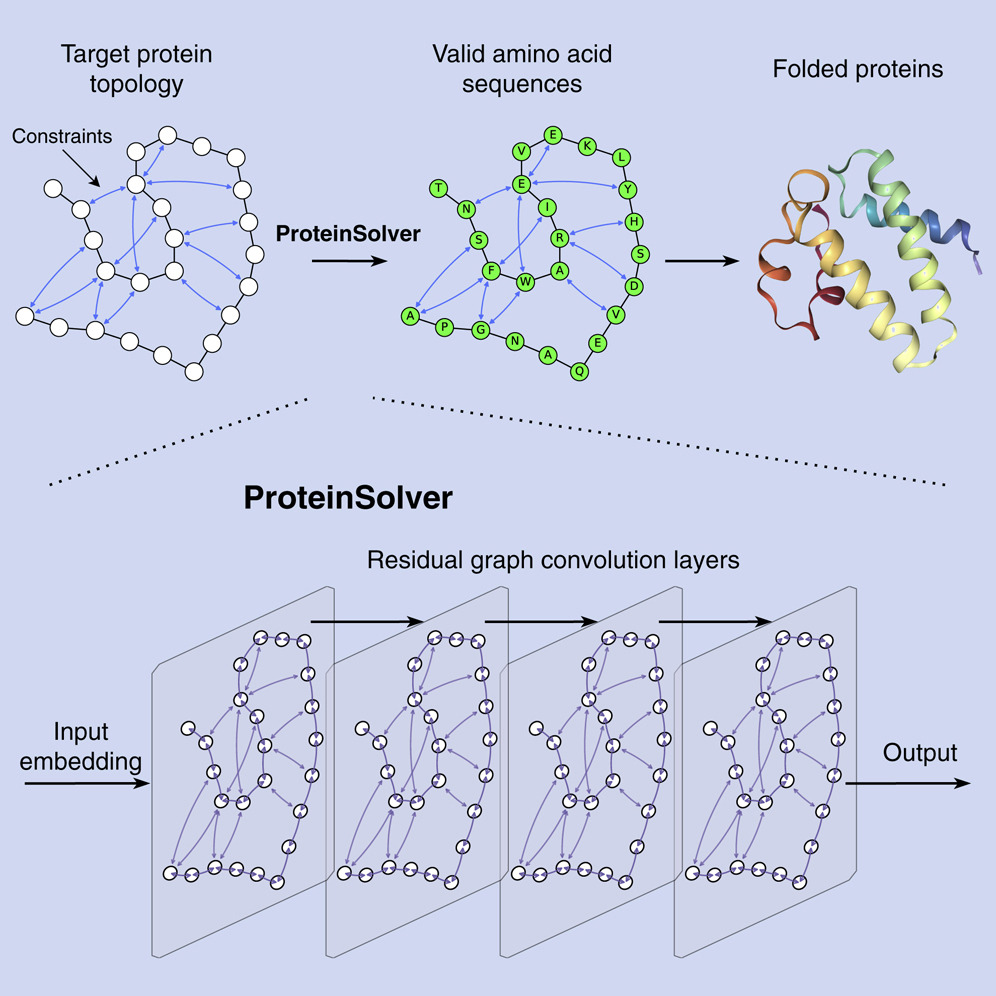
- Alexey Strokach*, David Becerra, Carles Corbi, Albert Perez-Riba and Philip M. Kim. Computational generation of proteins with predetermined three-dimensional shapes using ProteinSolver. Star protocols 12 (2), June 2021.
- Alexey Strokach*, David Becerra, Carles Corbi, Albert Perez-Riba and Philip M. Kim. Fast and flexible design of novel proteins using graph neural networks. Cell Systems 11 (4), 402-411, August 2020.
- Pedro Alberto Valiente*, David Becerra, Philip M. Kim. A method to calculate the relative binding free energy differences of α-helical stapled peptides. J Org Chem, 2020.
- David Becerra*, A. Butyaev, J. Waldispühl. Predicting Protein Folding Pathways using Ensemble Modelling and Evolutionary Sequence Variation. Oxford Bioinformatics, 2019, btz743.
- Zheng Dai*, David Becerra, J. Waldispühl. On Stable States in a Topologically Driven Protein Folding Model. Journal of Computational Biology, 2017, 24,9.
- David Becerra*, Juan Mendivelso, Yoan Pinzon. A Multiobjective Optimization Algorithm for the Weighted LCS, Journal of Discrete Applied Mathematics, 2015,11,005.
- Wilson Soto*, David Becerra*. A Multi-Objective Evolutionary Algorithm for Improving Multiple Sequence Alignments. Advances in Bioinformatics and Computational Biology, 73-82 (Brasil, 2015).
- D. Kwak*, A. Kam*, David Becerra*, Q. Zhou, A. Hops, E. Zarour, A. Kam, L. Sarmenta, J. Waldispühl. Open-Phylo: A customizable crowd-computing platform for multi sequence alignment. Genome Biology, 2013, 14:R116.
- Angelica Sandoval*, David Becerra*, Daniel Restrepo-Montoya, Diana Vanegas, Luis F. Nino. A Multi-objective Optimization Energy Approach to Predict the Ligand Conformation and Configuration in a Docking Process. European Conference on Evolutionary Computation, Machine Learning and Data Mining in Computational Biology, EvoStar 2013, (Vienna, Austria. 2013).
- David Becerra*, Juan Mendivelso, Yoan Pinzon. A Multiobjective Approach to the Weighted Longest Common Subsequence Problem. In Proceedings of the Prague Stringology Conference 2012 (PSC 2012), (Prague, Czech Republic. 2012).
- Daniel Restrepo-Montoya*, David Becerra*, Juan Guillermo Carvajal-Patino, Alvaro Mongui, Manuel Alfonso Patarroyo. Identification of Plasmodium vivax proteins with potential role in invasion using sequence redundancy reduction and profile hidden Markov models. PLoS ONE, 2011, 6(10).
- Dipankar Dasgupta*, Deon Garrett, Fernando Nino, Alex Banceanu and David Becerra*. A Genetic-Based Solution to the Task-Based Sailor Assignment Problem. Variants of Evolutionary Algorithms for Real-World Applications, Springer- Verlag 2011.
- David Becerra*, Angelica Sandoval, Daniel Restrepo-Montoya, Fernando Nino. A Parallel Multi-objective Ab-Initio Approach for Protein Structure Prediction. In Proceedings of the 2010 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2010), (Hong Kong, China, Dec 18 - 21, 2010).
- Dipankar Dasgupta*, David Becerra*, Alex Banceanu, Fernando Nino, James Simien. A Parallel Framework for Multi-objective Evolutionary Optimization. In Proceedings of the 2010 IEEE World Congress on Computational Intelligence (WCCI 2010), (Barcelona, Spain, Jul 18 - 23, 2010).
- David Becerra* and Wilson Soto, Fernando Nino, Yoan Pinzon (2010), An Algorithm for Constrained LCS, In Proceedings of the 8th ACS/IEEE International Conference on Computer Systems and Applications (AICCSA10), (Hammamet, Tunisia, 2010).
- David Becerra*, Diana Vanegas, Giovanni Cantor, Fernando Nino. (2009), An Association Rule based Approach for Biological Sequence Feature Classification, In Proceedings of the IEEE Congress on Evolutionary Computation (CEC 2009) (Trondheim, Norway, May 18 - 21, 2009).
- David Becerra* and Sergio Duarte, Fernando Nino, Yoan Pinzon, A Novel Image Encryption Scheme Based on a Generalized Chinese Remainder Theorem, Poster Presentation of the 2008 International Workshop on Combinatorial Algorithms (IWOCA 2008), (Nagoya, Japan. September 13-15, 2008).
- David Becerra* and Sergio Duarte*, Fernando Nino, Yoan Pinzon, A Novel Ab-Initio Genetic Based Approach for Protein Folding Prediction, In Proceedings of the 2007 Conference on Genetic and Evolutionary Computation (GECCO'07) (London, UK, July 7 - 11, 2007), pages 393-400, ACM Press.