
 Chromatin structure
Structural biology
Structure prediction
Chromatin structure
Structural biology
Structure prediction
Welcome! My name is Sol Shenker, and this page is intended to demonstrate a few of the research projects I have worked on during my undergraduate degree. I graduated in spring 2009 with a B.Sc. degree in Biochemistry and a minor in Computer Science. Currently, I am continuing my studies in Computer Science/Computational Biology at McGill.
The applet below is Chromoscope, a program I wrote in 2008 while working in Dr. Josee Dostie's. Chromoscope models chromatin structure based on the interaction frequency data acquired through 3C and 5C experiments. Interaction frequencies are transformed into distance constraints, and through gradient descent the program searches for models for which these constraints are satisfied. The goodness of fit of a model is represented by an energy score in the upper left hand corner, summarizing how well all the constraints are satisfied.
The program has been initialized with some mock data for a region of supercoiled chromatin. By pressing 'Start' the computation is initiated, and can be paused at any time using the 'Stop' button. The structures generated can be examined by dragging the mouse to rotate the model. The constraints for this mock data can be visualized by pressing the 'Constraints' button; red lines represent poorly satisfied constraints, and green lines satisfied constraints. Since gradient descent is prone to finding local minima, the 'Jitter' and 'Restart' buttons are provided. Jitter randomly pertubs each point randomly, while restart reinitializes the model with a random structure. Finally, if desired the model can also be manually perturbed in 'Drag' mode.
One of my projects in Dr. Kalle Gehring's lab was to determine the structure of the Ubiquitin-like domain of SACSIN. To do this I took a two pronged approach, simultaneously using NMR and crystallographic techniques. Unfortunately, the protein was poorly expressed in the minimal media necessary for the isotopic labeling, so I instead focused my efforts on crystallizing the protein. I successfully determined conditions in which the protein crystalized, and after optimization obtained crystals that diffracted to 2.2 angstroms on the home source X-ray beam.
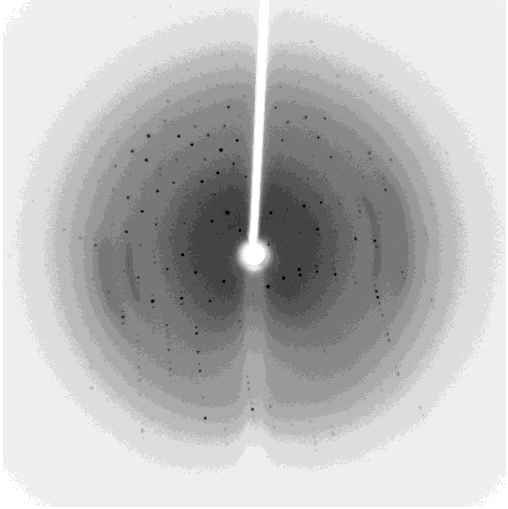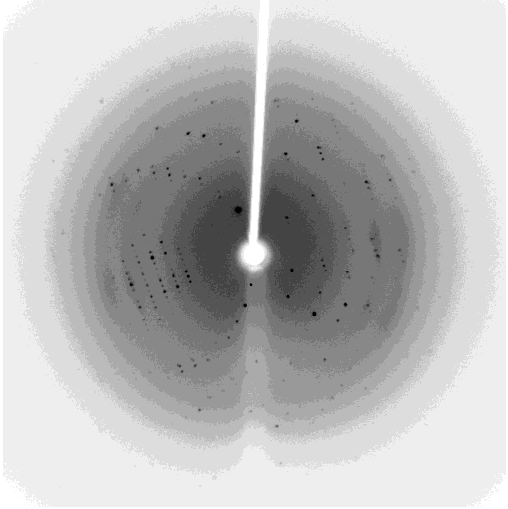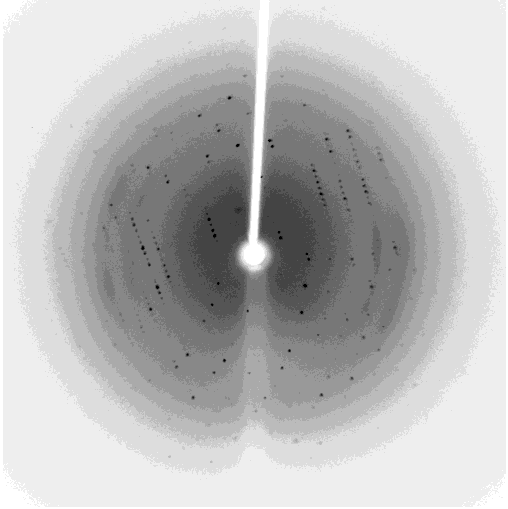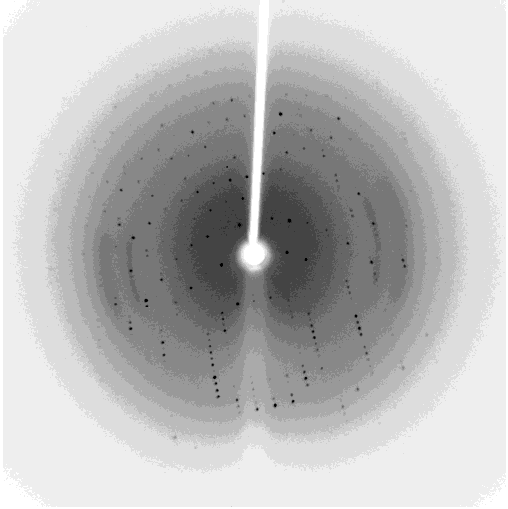
Indexing showed that the crystals had C2221 symmetry, without too much mosaicity. Since Ubiquitin-like domains share structural homology we logically attempted to use Ubiquitin as a model for molecular replacement. Suprisingly, Ubiquitin was not a suitable model for molecular replacement. Not to be so easily thwarted, I wrote a script that interfaced with Phaser to attempt molecular replacement with all the ubiquitin like structures in the protein databank. This too was unsuccessful, so as a final effort at molecular replacement I generated models for molecular replacement computationally. Ubiquitin-like domains a characterized by a structurally conserved beta-grasp fold. I generated a putative structure of SACSIN's Ubiquitin-like domain using protein threading, and by using the Modeller libraries I further explored the possible conformations by generating hundreds of randomly perturbed structures that maintained the beta-grasp fold.
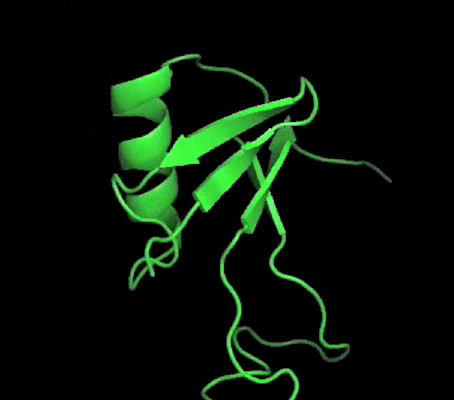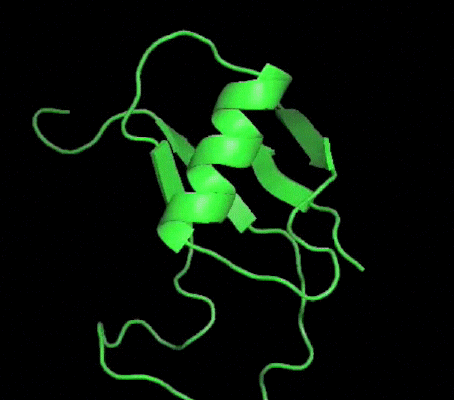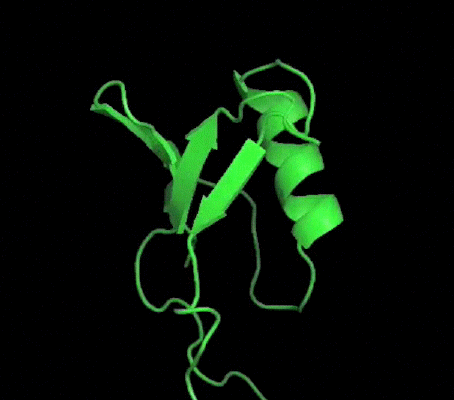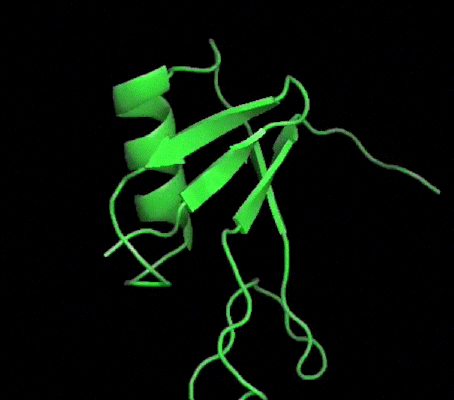

Unfortunately, this (informed) brute force approach to molecular proved unsuccessful. Efforts are continueing in the lab to generate selenomethionine labeled protein to solve the phase problem using multi-wavelength anomalous diffraction.