Tag set design algorithms, thermodynamic model, Affymetrix GenFlex, validation and verification
May 9, 2002
Prepared by Kaleigh Smith
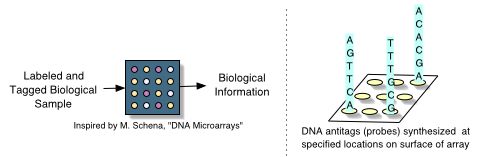
Universal DNA microarrays are organism-independent tools with the ability to recognize specific target genomic sequences or nucleic acid reaction products within a large and complex mixture. The microarray itself is a high-density matrix of oligonucleotides each designed to signal the presence or absence of a specific genomic sequence. An oligonucleotide is a short (20 - 25bp) sequence of amino acids, usually DNA though artificial bases are also used. When immobilized to the surface of a microarray, an oligonucleotide is named a probe, anti-tag or tag-probe. The Watson-Crick complement of a probe is a tag.
The basis of microarray technology is the hybridization between Watson-Crick complementary tag and probe pairs into stable duplex structures. Undesirable hybridization behaviours such an cross-hybridization, dimerization and secondary structure formation lead to inaccurate experimental results. Hybridization therefore introduces complications to the design of microarrays. The problem in designing microarrays is to select a maximally large set of oligonucleotide probes to be fixed to the surface of the array while strictly ensuring that each probe signals the presence or absence of its intended target with very high specificity. We will discuss issues with microarray design and several existing algorithms in further sections of this document.

Universal microarrays are a more flexible type of microarray than the popular organism-specific DNA microarrays. With organism-specific microarrays, the probes are designed from sections of target genomic sequences (usually genes). These probes are either immobilized on the surface of the array or are placed in wells on the array. DNA or RNA is isolated from a biological sample, processed into target sequence segments and then labeled to render them optically detectable. These targets are washed over the microarray (or chip) and hybridize with their complementary probes.
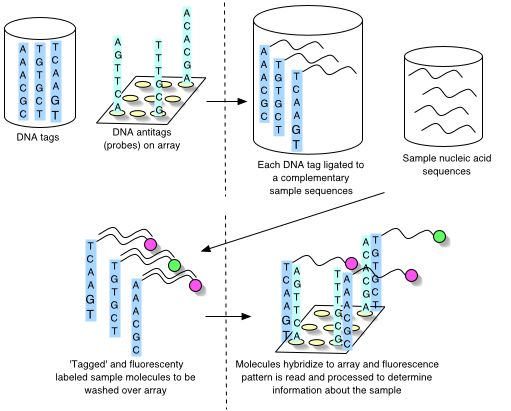
Universal microarrays differ because their probes are not targeted to an organism-specific genomic sequence or product. Instead, probe-complementary tags are ligated to the target-complementary sequences. These ligated molecules then hybridize with the tagged target sequences in the sample solution. There are then a variety of methods for fluorescently (or otherwise) labeling the tag-containing molecule [2,,,,,21]. The resulting tagged and labeled molecules are brought in contact with the universal microarray and are captured by each tag's complementary probe. The makeup of the sample can then be identified as the researcher knows which tag molecule hybridized to which target sequence.
These microarrays are used for SNP detection, genotyping of SNPs, gene expression profiling, and to facilitate custom design hybridization-based assays. Affymentrix writes of their commercial universal microarray GenFlex Tag Array, äny reaction in which a nucleic acid tag can be incorporated into the products may be amenable to analysis with the array"[1]. In each application, the universal microarray is used to determine the identity or abundance of target nucleic acid samples.
*For more details on the manufacture, data processing and quality control of organism-specific and universal microarrays, please see summany document by Fu Chen.*
A universal microarray can be represented by the set of unique probes immobilized on its surface, or equivalently, by the set of unique tags complementary to those probes. An oligonucleotide, and thus a tag and a probe, is commonly represented as a string of letters over the DNA alphabet. It is useful to abstract to these representations when considering the problem of designing a universal microarray. Such abstractions also help to define and measure hybridization behaviours of oligonucleotides.
Definition 1 [Tag] A tag t = < t1t2 ... tn > is an oligonucleotide (sequence of bases) over a DNA or
extended DNA alphabet with n between 15 and 25. The left to right ordering of a string
corresponds to 5' to 3' orientation of an oligonucleotide. In string notation, let Sdnan = { A, C, G, T }. A tag t of length n is a string t = t1 t2 ¼tn Î Sdnan.
Example:
t = ACTTGCAATCGTAATCGC = 5¢-ACTTGCAATCGTAATCGC-3¢.
The reverse tr of t is the string tn tn-1 ¼t1.
Definition 2 [Anti-tag] A complement tag, or anti-tag [`t] is the Watson-Crick complement of tag t. Each base of t is replaced by its Watson-Crick complement and the orientation of the derived sequence is reversed. In string notation, the Watson-Crick complement [`t] of t is the string obtained
from tr by interchanging A « T and C « G.
We say that two strings s and t are a Watson-Crick complement pair
iff [`s] = t.
Example: If t = 5¢-ACTTGCAATCGTAATCGC-3¢ then
[`t] = 5¢-GCGATTACGATTGCAAGT-3¢
5' - A-C-T-T-G-C-A-A-T-C-G-T-A-A-T-C-G-C - 3'
| | | | | | | | | | | | | | | | | |
3' - T-G-A-A-C-G-T-T-A-G-C-A-T-T-A-G-C-G - 5'
Self-Hybridization
A tag t may self-hybridize to form a secondary structure. As we model short DNA sequences, we will consider only the most simple folded structure, the hairpin loop. In string notation, a tag t self-hybridizes to form a perfect hairpin structure with a loop of size k and a stem of length l iff ttop = [`(tbottom)] where ttop = t1 t2 ¼tl and tbottom = ts+k+1 ts+k+2 ¼tn.An example, t = AAAAAAAAGGGTTTTTTTT has the potential to form a stable hairpin structure:
5' - A-A-A-A-A-A-A-A-G
| | | | | | | | G
3' - T-T-T-T-T-T-T-T-G
Self-Dimerization A tag t perfectly self-dimerizes, that is two copies of t hybridize perfectly iff tr = [`t]. For instance, if two copies of t = TTTCCCATGGGAAA are present in a solution, they will hybridize to form a stable duplex:
5' - T-T-T-C-C-C-A-T-G-G-G-A-A-A - 3'
| | | | | | | | | | | | | |
3' - A-A-A-G-G-G-T-A-C-C-C-T-T-T - 5'
Imperfect Hybridization
Two tags s and t hybridize imperfectly iff s » t. For instance, the edit distance between s and t may be very small, or s and t may contain a long common substring.
Cross-Hybridization Tags t and s exhibit cross-hybridization if t » s, so that t hybridizes imperfectly to [`s] and s hybridizes imperfectly to [`t]. Consider s = TTACCGTTAAGTAC, t = TTACCCTTAAGTAC and [`t] = GTACTTAAGGGTAA. s will hybridize imperfectly to [`t] (and t to [`s]):
5' - T-T-A-C-C-G-T-T-A-A-G-T-A-C - 3'
| | | | | | | | | | | | |
3' - A-A-T-G-G-G-A-A-T-T-C-A-T-G - 5'
Dimerization Tags t and s exhibit dimerization if t » [`s], so that t and s hybridize imperfectly. Consider s = TTACCGTTAAGTAC and t = GTACTTTAACGATAA:
5' - T-T-A-C-C-G-T-T-A-A-G-T-A-C - 3'
| | | | | | | | | | | |
3' - A-A-T-A-G-C-A-A-T-T-T-A-T-G - 5'
For a set of tags T = { t1, ¼, tm }, we use [`T] to denote the set of anti-tags obtained from T: [`T] = { [`(ti)] : 1 £ i £ m }. A specific microarray chip C is represented by an ordered pair C = áT, Añ where T is a set of tags { t1, ¼, tm }, A is a set of tags { a1, ¼, an }, ti Î Sdnan, ai Î Sdnan and there is a bijection s: T ® A s.t. [`t] = s(t). That is, every tag t Î T has exactly one Watson-Crick complement s(t) = a Î A, so m = n.
Definition 3 [Good Tag Set]
A good tag set is set of tags T such that

A combinatorial model for DNA hybridization behaviour simply counts the number of base pairs that form Watson-Crick pairs and those that mismatch. Such a model is not based on the biochemical properties that govern the hybridizations, but is rather based on the similarity of the two tags in question. Such a model commonly uses measurements of distance between strings to predict the occurrence of imperfect hybridizations (Hamming distance, longest common substring or subsequence).
Several models have been proposed in the literature to analyze DNA hybridization by estimating thermodynamic properties governing the formation of a duplex. Thus, hybridization behaviour corresponds to duplex stability and is popularly measured in terms of melting temperature or free energy. The melting temperature TM of two tags is the temperature at which half of each strand's nucleotides are in double-helical duplex form and half are in single-stranded form. The most simple model for estimating the TM of a duplex is the 2-4 model consisting of a simple rule based on the number of hydrogen bonds between a two nucleotides. With the 2-4 model, the TM of a sequence and its Watson-Crick complement is
TM = 2 (number of A-T base pairs) + 4(number of C-G base pairs) [19].
A thermodynamic nearest-neighbour model of DNA hybridization offers a more specific and biologically based model for short DNA sequences. Several sets of NN base pair parameters have been introduced, and we use the most well known and accurate parameters determined by SantaLucia [14]. Though this model can be applied to predict the TM of two tags, we focus on its ability to predict the free energy, dG of a duplex. dG has been shown to be more accurate and robust to errors than the dH and dS properties that are used to determine TM [14].
We have seen that two very similar tags will cross-hybridize to each other's anti-tags. It is very likely that such tags will share a long common substring, though there may be a possibility for cross-hybridization when there are many shorter common substrings and weakly mismatched base pairs. Still, long common substrings are a satisfactory indicator of cross-hybridization. The problem of good tag set design can then be to find the largest possible set of tags while ensuring that no two tags are overly similar. As described in [4], this can be accomplished by constructing the largest possible l-free code. A set of strings is a l-free code iff no two strings in the set have a common substring of length greater than l.
Input: Given integers n, l and an alphabet of bases S.
Output: A maximally large set T of length n strings over S such that no two strings s, t Î T have a common substring of length greater than l.
This problem can be solved by parsing a de Bruijn sequence of order l to construct an optimal l-free code of size 4l/(n - l+ 1) [18].
Definition 4 [De Bruijn sequence] The shortest circular sequence of length l|S| such that every string of length l on the alphabet S of size |S| occurs as a contiguous subrange of the sequence described by S. For example, a de Bruijn sequence of order 2 on the alphabet {a, b, c} is given by {aacbbccab}. [22]
This approach can be extended to create a tag set that avoids dimerization. This is accomplished by further filtering the constructed code T to ensure that for all tags t Î T, there is no tag s Î T such that t contains a substring x and s contains the x's complement [`x]. It is also clear that another filter may be applied to T to remove any tags that contain both substring x and the reverse of [`x], as such a tag would likely exhibit self-dimerization.
Input: Given integer n, value t and an alphabet of bases S.
Output: A maximally large set T of length n strings over S such that no two strings s, t Î T form a duplex with thermodynamic stability measurement greater than t.
A greedy algorithm to solve this problem would begin with an empty set of tags T and add a new tag, randomly chosen or chosen according to some heuristic, iff it is not predicted to cross-hybridize, dimerize or form self-structures. A greedy strategy for avoiding cross-hybridization is described in [18]. Unfortunately, a greedy approach does not ensure a maximally large set of tags and additional constraints would be necessary to bias the algorithm towards adding tags that form strong duplexes at similar temperatures.
Affymetrix's approach to universal microarray technology includes adding additional mismatch probes (one base pair different from an actual probe) without complementary tags to the array to measure the specificity of each 'signalling' or actual probe. Affymetrix also includes control probes that are the complements of each actual probe and mismatch probe to detect possible dimerization. The critical observer will notice that the naive tag design algorithm necessitates the additional mismatch and control probes to identify and subtract unwanted reactions such as cross-hybridization and dimerization.
Good Individual Tags
Each tag selected for the final tag set must be guaranteed to hybridize with its complementary anti-tag when the experiment is performed at some given temperature t. This is done by designating temperature thresholds C and H, C < t < H. For each tag t in the set of good tags T, tM(t, [`t]) ³ H.
Good Pairwise Tags
Just as each intended hybridization must occur for the tag set to be good, no unintended hybridizations can be allowed to occur. Using the same temperature thresholds C and H as described above, for each pair of tags t and s in the set of good tags T, tM(s, [`t]) < C (and tM(t, [`s]) < C). The authors then make the well-supported assumption that tM(s, [`t]) < C (and tM(t, [`s]) < C) occurs iff t and s have a common substring x and tM(x, [`x]) ³ C.
It follows that the good tag set design problem defined in [3] is:
Input: Given threshold values C and H and an alphabet of bases S.
Output: A maximally large set T of tags over S such that:
A string s has weight w(s), w(s) = 1/2(tM(s)) when tM is calculated according to the 2-4 rule. So, with respect to the previous definitions, 2c = C and 2h = H.
Definition 5 [c-h code] A tag set T is a valid c-h code iff each tag t Î T has w(t) ³ h and and every substring of every tag t in T of weight c or more occurs in no other tags t¢ Î T, t ¹ t¢ and occurs at most once in t. [3]
The authors of [3] define the Combinatorial Tag Design Problem as the problem of constructing a maximally large c-h code for appropriate parameters c and h. The authors then combine de bruijn sequences and novel combinatorial approaches to create an algorithm to generate an optimal c-h code and to provide bounds on the size of T.
Many such quality control algorithms proceed by introducing additional probes onto the array. Hubbell and Pevzner in [12] describe an idea to add identical control probes in several locations over the microarray. These control probes must be immobilized to the microarray in different ways, using as many different manufacturing steps as possible. If there is no manufacturing error at all of these steps, all control probes are expected to display near-identical intensity readings. However, if one control probe is incorrectly created, its intensity will be dissimilar. It is this principle that allows control probes to detect manufacturing error in microarrays. It may strengthen the approach to have several different control probes, as this increases the number of steps in the manufacturing process that may be monitored[16].
The problem now becomes the design of such control probes. The authors of [16] define a problem similar to that of universal microarray tag design. In addition to selecting a set of control probes that have good tag set properties among their set, the control probes must also maintain good tag set properties when combined with the set of experimental tags. The problem is further complicated by the need to ensure that some non-zero number of probes participate in every manufacturing step of the microarray. A solution to this problem, and a more complete description of the issues arising from microarray verification can be found in [16].