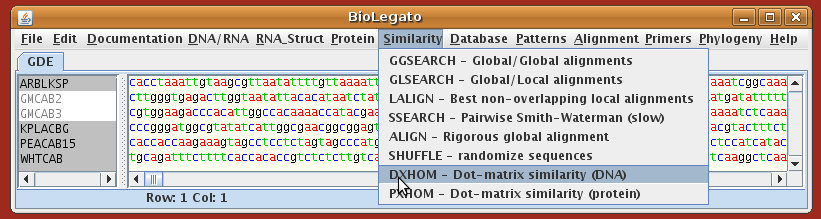
DXHOM documentation: $doc/fsap/hom.asc
total 58
-rw------- 1 frist drr 5404 Nov 6 17:22 ARBLKSP.gen
-rw------- 1 frist drr 4494 Nov 6 17:21 GMCAB2.gen
-rw------- 1 frist drr 4238 Nov 6 17:21 GMCAB3.gen
-rw------- 1 frist drr 4338 Nov 6 17:29 KPLACBG.gen
-rw------- 1 frist drr 3674 Nov 6 17:21 PEACAB15.gen
-rw------- 1 frist drr 3757 Nov 6 17:22 WHTCAB.gen
Read in the GenBank files using File --> Open.
Hint: Similarity
comparisons require you to select two or more sequences at a time.
|

Select GMCAB2 and GMCAB3 by dragging the cursor across the two names, and choose Similarity --> DXHOM. The DXHOM menu will appear:
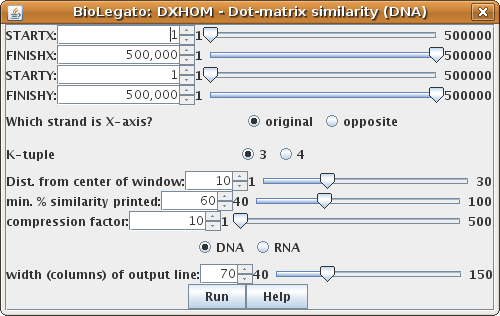
Output from GMCAB2 vs. GMCAB3 using
DXHOM defaults.
a. Compression: Zooming in and out - Each row and column in the matrix represents one or more nucleotide/amino acid positions. By default, A compression factor of 10 is used, with 70 charcters per line, meaning that 700 nucleotides can be represented per line or column. In the example above, the X-axis sequence GMCAB2 is 1354 bp long, so DXHOM must break the output up into two pages, one in which GMCAB2 positons 1..700 are comared with all of GMCAB3, and another in which 701..1354 are compared with GMCAB3. The entire comparison can be fit into a single page by changing the compression factor to 20. [Output with "compression factor" (COMPRESS) =20].b. The signal to noise ratio is controlled by local search window size and similarity cut off value.
The parameter "min. % similarity printed (MINPER)" sets the minimum score for a character to be printed in the matrix. The lower the percent match allowed, the higher the sensitivity, but the greater the background noise due to random chance. [Output with min. % similarity printed=50].
The window size is expressed as the distance from the center of the search window. That is , the search window contains d bases on either side of the center of the k-tuple match. So if the distance is 10, a window of 21 bases (ie. 10 on either side of the central nucleotide) is compared. The wider the search window, the lower the probability of a match by random chance. Thus, lower MINPER values require larger search window sizes. The result from above can be "cleaned up" by doubling the window size. [Output with window Dist. from center of window= 20 ie. window size = 41].
Some reduction in background noise can also be gained by using a k-tuple of 4. This will also speed up the search by a factor of 4, but may be less sensitive.
Finally, the signal to noise ratio can also be improved by simply doing the plot at a lower compression. One way is to use a wider output line (scroll left to right to see entire matrix)
Dist. from center of window: 10
MINPER = 50
COMPRESS = 10
width of output line: (LINEWIDTH) = 130
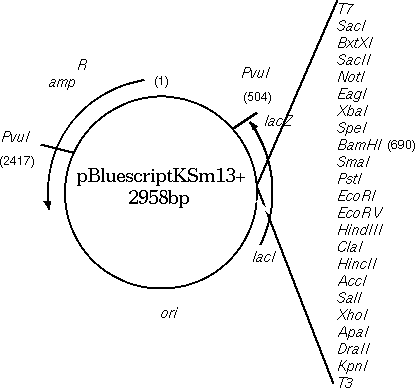
To illustrate the importance of comparing BOTH strands, the
Bluescript vector, containing a beta-lactamase gene for ampicillin
resistance, will
be compared with the beta lactamase gene from Klebsiella pneumonia.
Since Bluescript is 2958 bp long, we need to use COMPRESS= 30 and
LINEWIDTH=
100 to make the entire matrix fit onto one page.
| NOTE: If you plan
to compare both strands of a reference sequence with one strand
of another sequence, in each plot the reference sequence, or its
inverse complement, MUST be
placed on the X-axis.
DXHOM is designed to preserve the coordinate system of the
reference sequence, regardless of which strand is printed. That is, if
you do the
first search with the original strand Bluescript as the reference
sequence,
the coordinates of Bluescript would be written left to right on the
X-axis,
1 to 2958. It is INCORRECT to simply generate the inverse complement of
Bluescript and repeat the search, numbering 1.. 2958. The numbering at
the top should be 2958 downto 1. In that way, a given coordinate always
refers to the same part of the sequence, regardless of the strand. |
Although there's no straightforward way to get biolegato to tell DXHOM which sequence goes onto the X-axis, when multiple sequences are selected, they are sent to programs in the order they appear in the biolegato sequence list, top to bottom. Thus, if you want ARBLKSP to be on the X-axis, it must appear above KPLACBG in the biolegato window.
Compare the original strand of Bluescript (ARBLKSP) with the Klebsiella gene (KPLACBG).
Dist. from center of window: 10As seen in the output, there are no significant diagonals, indicating that there are no
min % similarity: 60
COMPRESS=30
LINEWIDTH=100
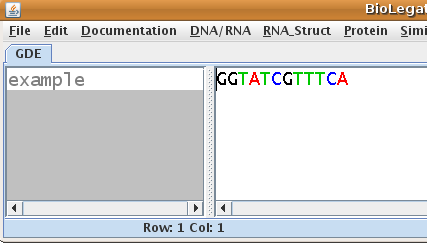
| As shown at right, example
is written 5' to 3' left to right. Select example, and
choose DNA/RNA --> blrevcomp.
For any sequence, blrevcomp can create the complementary strand, or
flip the strand, or create the inverse-complement. |
 |
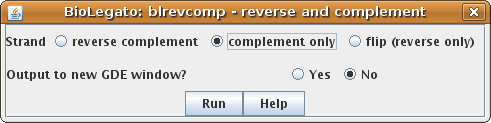
| Let's start by creating the
complementary strand. Choose complement
only. |
 |
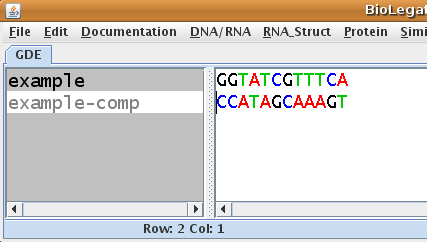
| Since
example is written 5' to 3' left to right, it should be obvious that
example-comp as displayed in the figure, must be written 3' to 5', left
to right. (Note that blrevcomp automatically adds a suffix to the sequence name to distinguish between the original strand and its diferent transformations.) Next, select example-comp and run blrevcomp again, this time choosing flip (reverse only). |
 |
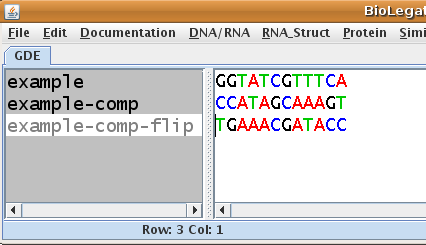
| Now, example-comp-flip is the
same strand as example-comp, but written 5' to 3' left to right. |
 |
| blrevcomp can achieve the same
result in one step if you select example and choose reverse-complement in the blrevcomp
menu. As we'd predict, example-opp is identical to example-comp-flip. |
 |
a. Create the inverse complement of ARBLKSP.
Select ARBLKSP and choose DNARNA
--> blrevcomp.
The inverse complement, ARBLKSP-opp, is inserted at the bottom of
the biolegato window.
When biolegato runs DXHOM, it places top-most sequence on the X axis
and the lower sequence on the Y axis. DXHOM is written in such a way
that it can recalculate the coordinates of the X-axis sequence if that
sequence is an inverse complement. For that reason, we need to move
ARBLKSP-opp up in the biolegato window.
Select ARBLKSP-opp and choose Edit
--> Cut. To insert it below ARBLKSP, click on 'ARBLKSP'
and choose Edit --> Paste.
b. Compare the inverse-complement of ARBLKSP with KPNLACBG.
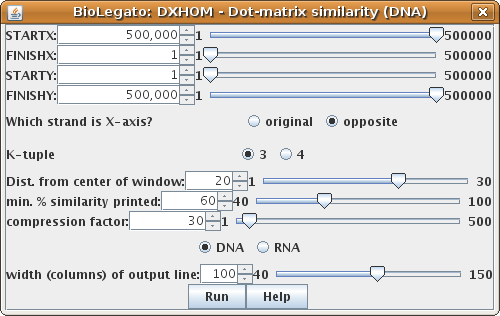
Select ARLKSP-opp and KPNLACBG. Choose DNA/RNA --> DXHOM.| Click on the opposite button to tell DXHOM that
the X-axis strand is the opposite strand. That will tell DXHOM to write the sequence coordinates in
descending order. This also requires that we start (STARTX) at
the end of the sequence and finish (FINISHX) at the begining of the
sequence. Drag the STARTX slider to the right, and the FINISHX slider
to the left. If you don't do this step, the sequence position numbers in the output will be incorrect! |
 |
As illustrated in the map of Bluescript shown above, the
DXHOM output verifies that the opposite strand of Bluescript has a
beta-lactamase gene, going roughly from 2680 downto 1960. [DXHOM output using opposite strand
of ARBLKSP].